Publicações
Relógio Biológico de Plantas
Hotta CT (2022) The evolution and function of the PSEUDO RESPONSE REGULATOR gene family in the plant circadian clock. Genet Mol Biol. 45 (3 Suppl 1)
Ribeiro C, Stitt M, Hotta CT (2022) How stress affects your budget – Stress impacts in starch metabolism. Frontiers in Plant Science
Dantas LLB, de Lima NO, Cavaçana N, Dourado MM, Carneiro MS, Souza GM, Caldana C, Hotta CT (2021) Microenvironments regulate diel rhythms in the field. New Phytologist 232(4)
Hotta CT. From Crops to Shops: How Agriculture Can Use Clocks (2021) Journal of Experimental Botany 72(22): 7668-79
Dantas LLB, Almeida-Jesus FM, de Lima N, Alves-Lima C, Nishiyama-Jr. MY, Carneiro MS, Souza GM, Hotta CT (2020) Rhythms of Transcription in Field-Grown Sugarcane Are Highly Organ Specific. Sci Rep 10: 6565
Dantas LLB, Calixto CPG, Carneiro MS, Brown JWS, Hotta CT (2019) Alternative splicing of circadian clock genes is temperature regulated in field-grown sugarcane. Front Plant Sci 10:1614.
Mombaerts L, Carignano A, Robertson FR, Hearn TJ, Junyang J, Hayden D, Rutterford Z, Hotta CT, Hubbard KE, Martí Ruiz MC, Yuan Y, Hannah MA, Gonçalves J, Webb AA (2019) Dynamical Differential Expression (DyDE) Reveals the Period Control Mechanisms of the Arabidopsis Circadian Oscillator. PLoS Comput Biol 15(1): e1006674
Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE, Wells Newman D, Yochikawa A, Cano-Ramirez DL, Chembath A, Cragg-Barber K, Haydon MJ, Hotta CT, Vincentz M, Webb AAR, Dodd AN (2018) Circadian Entrainment in Arabidopsis by the Sugar-Responsive Transcription Factor bZIP63. Curr Biol 28(16):2597-2606.e6
Martí Ruiz MC, Hubbard KE, Gardner MJ, Jung HJ, Aubry S, Hotta CT, Mohd-Noh NI, Robertson FC, Hearn TJ, Tsai YC, Dodd AN, Hannah M, Carré IA, Davies JM, Braam J, Webb AAR (2018) Circadian oscillations of cytosolic free calcium regulate the Arabidopsis circadian clock. Nat Plants. 690-698.
Hotta CT, Nishiyama MY Jr, Souza GM (2013) Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PLoS One 8(8):e71847. [link] [pdf]
Dalchau N, Hubbard KE, Robertson FC, Hotta CT, Briggs HM, Stan GB, Gonçalves JM, Webb AA. (2010) Correct biological timing in Arabidopsis requires multiple light-signaling pathways. PNAS 107(29):13171-6. [link] [pdf]
Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Robertson FC, Love J, Sanders D, Webb AA. (2009) Response to Comment on “The Arabidopsis Circadian Clock Incorporates a cADPR-Based Feedback Loop”. Science 326, 230-1. [link] [pdf]
Hotta CT, Xu X, Xie Q, Dodd AN, Johnson CH, Webb AA. (2008) Are there multiple circadian clocks in plants? Plant Signal Behav. 3, 342-4. [link] [pdf]
Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Gonçalves J, Sanders D, Webb AA. (2007) The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318, 1789-92. [link] [pdf]
Xu X, Hotta CT, Dodd AN, Love J, Sharrock R, Lee YW, Xie Q, Johnson CH, Webb AA. (2007) Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell. 19, 3474-90. [link] [pdf]
Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AA. (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 30, 333-49.[link] [pdf]
Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AA. (2006) How plants tell the time. Biochem J. 397,15-24. [link] [pdf]
Biologia da cana-de-açúcar
Souza GM, Sluys MAV, Lembke CG, Lee H, Margarido GRA, Hotta CT, Gaiarsa JW, Diniz AL, Oliveira MM, Ferreira SS, Nishiyama-Jr MY, ten Caten F, Ragagnin GT, Andrade PM, de Souza RF, Nicastro GG, Pandya R, Kim , Durham AM, Carneiro MS, Zhang J, Zhang X, Zhang Q, Ming R, Schatz VC, Davidson B, Paterson AH, Heckerman D (2019) Copy-resolved assembly of the sugarcane gene space reveals reservoirs of homo(eo)logs diversity. GigaScience, 8(12), giz129.
Alessio VM, Cavaçana N, Dantas LLB, Lee N, Hotta CT, Imaizumi T, Menossi M (2018) The FBH family of bHLH transcription factors controls ACC synthase expression in sugarcane. J Exp Bot. 69(10):2511-2525
Ferreira SS, Hotta CT, Poelking VG, Leite DC, Buckeridge MS, Loureiro ME, Barbosa MH, Carneiro MS, Souza GM (2016) Co-expression network analysis reveals transcription factors associated to cell wall biosynthesis in sugarcane. Plant Mol. Biol. 91(1): 15-35
de Setta N, Monteiro-Vitorello CB, Metcalfe CJ, Cruz GM, Del Bem LE, Vicentini R, Nogueira FT, Campos RA, Nunes SL, Turrini PC, Vieira AP, Ochoa Cruz EA, Corrêa TC, Hotta CT, de Mello Varani A, Vautrin S, da Trindade AS, de Mendonça Vilela M, Lembke CG, Sato PM, de Andrade RF, Nishiyama MY Jr, Cardoso-Silva CB, Scortecci KC, Garcia AA, Carneiro MS, Kim C, Paterson AH, Bergès H, D'Hont A, de Souza AP, Souza GM, Vincentz M, Kitajima JP, Van Sluys MA. (2014) Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics, 540
Lembke CG, Nishiyama MY Jr, Ferreira SS, Dal-Bianco ML, Sato PM, Hotta CT, Carneiro M, Loureiro M, Carrer H, Endres L, Hoffmann H, Sanches M, Menossi M, Souza GM (2013) Sugarcane genome sequencing and gene discovery: Getting closer to sugar content, fibre and drought traits. Cooperative Sugar 45(3), 23-27
Dal-Bianco M, Carneiro MS, Hotta CT, Chapola RG, Hoffmann HP, Garcia AA, Souza GM (2012) Sugarcane improvement: how far can we go? Curr Opin Biotechnol. 23(2):265-70. doi: 10.1016/j.copbio.2011.09.002.
Nishiyama-Junior MY, Vicente FFR, Lembke CG, Sato PM, Dal-Bianco M, Fandiño RA, Hotta CT, Souza GM (2012) The SUCEST-FUN regulatory network database: Designing an energy grass. International Sugar Journal. v. 114, p. 821
Hotta CT, Lembke CG, Domingues DS, Ochoa EA, Cruz GMQ, Melotto-Passarin DM, Marconi TG, Santos MO, Mollinari M, Margarido GRA, Crivellari AC, dos Santos WD, de Souza AP, Hoshino AA, Carrer H, Souza AP, Garcia AAF, Buckeridge MS, Menossi M, Van Sluys MA, Souza GM (2010) The Biotechnology Roadmap for Sugarcane Improvement. Tropical Plant Biology 3(2): 75-87
Casu RE, Hotta CT, Souza GM (2010) Functional Genomics: Transcriptomics of Sugarcane - Current Status and Future Prospects. In: Henry RJ; Kole C. (Org.). Genetics, Genomics and Breeding of Sugarcane. Routledge. Book chapter.
Outros
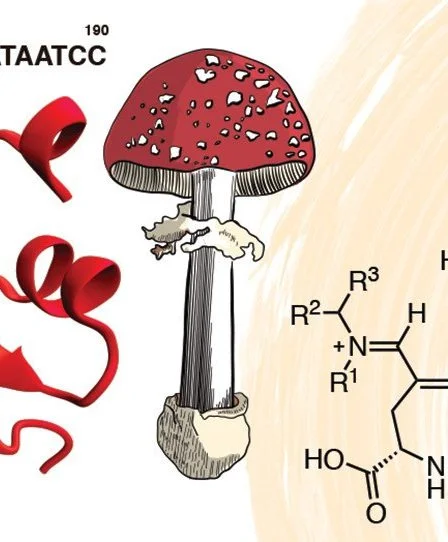
Soares DMM, Gonçalves LCP, Machado CO, Esteves LC, Stevani CV, Oliveira CC, Dörr FA, Pinto E, Adachi FMM, Hotta CT*, Bastos EL* (2022) Reannotation of Fly Amanita l-DOPA Dioxygenase Gene Enables Its Cloning and Heterologous Expression. *joint corresponding authors. ACS Omega
Baptista MS, Alves MJM, Arantes GM, Armelin HA, Augusto O, Baldini RL, Basseres DS, Bechara EJH, Bruni-Cardoso A, Chaimovich H, Colepicolo Neto P, Colli W, Cuccovia IM, da Silva AM, Di Mascio P, Farah SC, Ferreira C, Forti FL, Giordano RJ, Gomes SL, Gueiros Filho FJ, Hoch NC, Hotta CT, Labriola L, Lameu C, Machini MT, Malnic B, Marana SR, Medeiros MHG, Meotti FC, Miyamoto S, Oliveira CC, Pinto NCS, Reis EM, Ronsein GE, Salinas RK, Schechtman D, Schreier S, Setubal JC, Sogayar MC, Souza GM, Terra WR, Truzzi DR, Ulrich AH, Verjovski-Almeida S, Winck FV, Zingales BS, Kowaltowski AJ (2019) A Position Paper on Scientific Communication in Biochemistry and Molecular Biology. Braz J Med Biol Res 52(9): e8935
Alves-Lima C., Teixeira ARA, Hotta CT, Colepicolo P (2019) A cheap and sensitive method for imaging Gracilaria (Rhodophyta, Gracilariales) growth. J App Phycol 31(2): 885-892
Alves-Lima C., Cavaçana N., Chaves GAT, de Lima NO, Stefanello E, Colepicolo P, Hotta CT (2017) Reference genes for transcript quantification in Gracilaria tenuistipitata under drought stress. J. App. Phycol. 2017 29: 731-740
Abdul-Awal SM, Hotta CT, Davey MP, Dodd AN, Smith AG, Webb AA (2016) NO-Mediated [Ca2+]cyt Increases Depend on ADP-Ribosyl Cyclase Activity in Arabidopsis. Plant Physiol. 171(1):623-31
Hotta CT (2012) Sinalização de Cálcio em Plantas. In: Rodrigo Resende; Silvia Guatimosim. (Org.). Sinalização de Cálcio - Bioquímica e Fisiologia Celulares (Calcium Signalling - Biochemistry and Cell Physiology) 1ed. São Paulo: Editora Sarvier, p. 478-495. Book chapter
Hotta CT, Bandoni FO (2011) O que ensinar em Biologia? As pressões sentidas pelo professor ao selecionar os conteúdos (What to teach in Biology? The pressures felt by the teachers when selecting content). In: Rita de Cássia Frenedozo. (Org.). Pesquisa em ensino de ciências e biologia. 1ed. São Paulo: Terracota, p. 11-23. Book chapter
Bagnaresi P, Markus RP, Hotta CT, Pozzan T, Garcia CRS (2008) Desynchronizing Plasmodium cell cycle increases chloroquine protection at suboptimal doses. Open Parasitol J 2: 55-58
Hotta CT, Markus RP, Garcia CR (2003) Melatonin and N-acetyl-serotonin cross the red blood cell membrane and evoke calcium mobilisation in malarial parasites. Braz J Med Biol Res 36(11):1583-7
Hotta CT, Gazarini ML, Beraldo FH, Varotti FP, Lopes C, Markus RP, Pozzan T, Garcia CR (2000) Calcium-dependent modulation by melatonin of the circadian rhythm in malarial parasites. Nat Cell Biol 2(7):466-8